Multiscale Modelingmolecluar modeling
Coarse-Grained Molecaluar Model (SPICA force field)
![]()
We have developed a coarse-grained molecular force field (SPICA force field;
formerly called SDK (Shinoda-DeVane-Klein) force field) for surfactants
and lipids using multi-property fitting for thermodynamic quantities (particularly
related to interfacial properties like surface tension and molecular partitioning)
as well as distribution functions obtained from all atomic molecular dynamics
trajectories. Now the SPICA force field is extended to include carbon materials,
nanoparticles, peptides, proteins, DNA and RNA. We investigate soft materials
and biological molecular assembly using CG-MD with the SPICA force field.
Further details of the SPICA force field is provided from the SPICA-ff webpage. One can download parameter and topology files, several tools to run a
CG-MD using the SPICA force field.
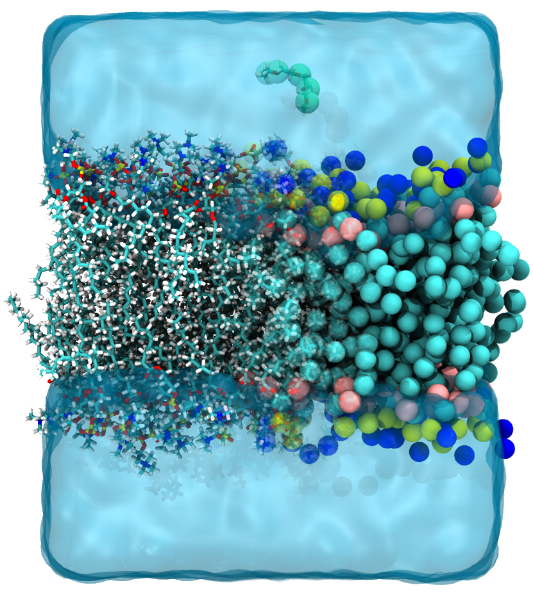 |
| The overlayed snapshot of a lipid bilayer system in AA and CG descriptions. |
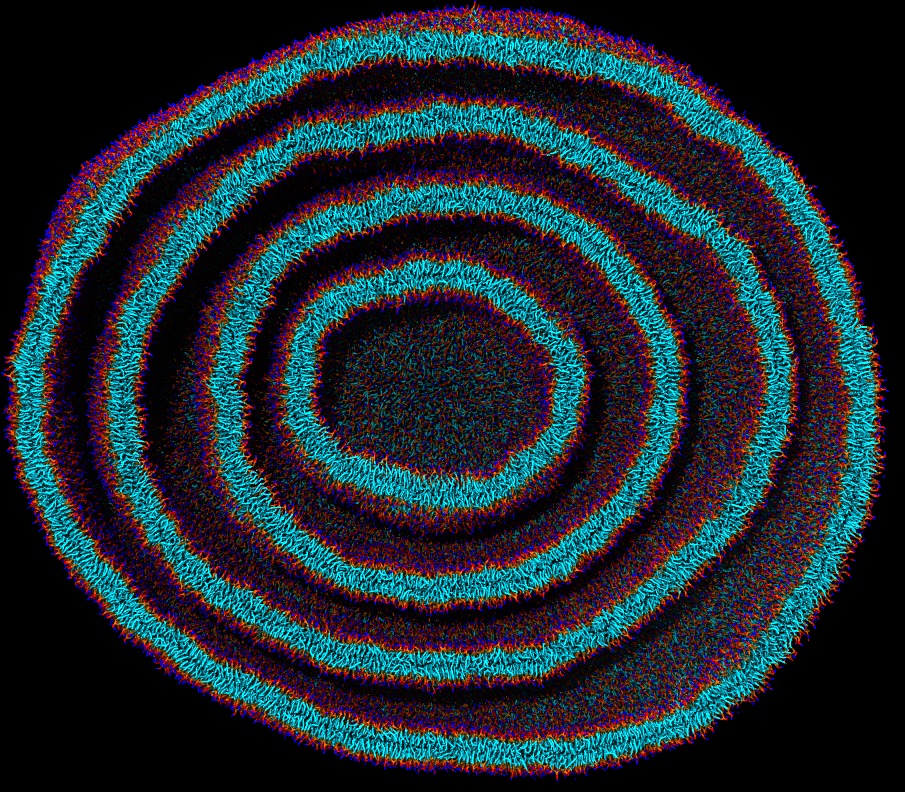 |
| Multi Lamellar Vesicle (MLV) made started from a random lipid aggregates in CG-MD with SPICA FF. |
Polarizable force field
Conventional force field used in molecular dynamics simulations for bio- and organic molecular systems usually consider a point chage on each atom on top of the Lennard-Jones nonbond interactions among atoms. However, for highly charged systems and their mixture with organic solvents like in ionic liquids and solvated lithium salts, consideration of induced polarizability is required to accurately describe the intermolecular interaction. We are developing a many-body polarizable force field on the basis of isotropic atomic induced dipole model especially for molecular dynamics simulation of solvated ionic liquids and solvated lithium salts systems, which are great candidate of the electrolyte in Li-ion, Li-sulfer batteries.


